This is the multi-page printable view of this section. Click here to print.
News about Virtual Fly Brain (VFB)
- 1: VFB workshop: 'Bridging connectomics and transcriptomics' at NeuroFly 2024
- 2: Guide to Working with Images from Virtual Fly Brain (VFB) Using the VFBConnect Library
- 3: Predicted neurotransmitters for EM data now available on VFB
- 4: New Split-GAL4 report pages on FlyBase
- 5: Transcriptomics data available for cell types on VFB
- 6: Larval split-GAL4 line raw images release
- 7: Fix for problems with current Safari release
- 8: Google search results issue
1 - VFB workshop: 'Bridging connectomics and transcriptomics' at NeuroFly 2024
The Drosophila neuroscience community now has access to extensive connectomics data covering the majority of the fly brain, along with a growing collection of transcriptomics data for brain cell types. Virtual Fly Brain integrates Drosophila neuroscience data, providing a platform for bridging these two data types and making them accessible for display, querying, and download.
The workshop will introduce the VFB API through practical examples, including querying and retrieving connectomics and transcriptomics data and integrating these data types in custom queries. This session is designed for experienced users of R or Python.
Thank you to everyone who attended our workshop at NeuroFly 2024. Our workshop notebook is available here: tinyurl.com/workshopVFB for anyone who would like to explore the VFB API. We will keep this available and up to date.
2 - Guide to Working with Images from Virtual Fly Brain (VFB) Using the VFBConnect Library
Prerequisites
Before starting, ensure you have the VFBConnect library installed. The recommended Python version is 3.10.14, as this version is tested against the library.
pip install vfb-connect
Importing the VFBConnect Library
Start by importing the VFBConnect library. This library provides a simple interface to interact with neuron data from the Virtual Fly Brain.
from vfb_connect import vfb
Retrieving Neuron Data
To work with specific neurons, you can use the vfb.term() function. This function takes a unique identifier (e.g., ID, label, synonym) for the neuron.
Example: Retrieving a Single Neuron
neuron = vfb.term('5th s-LNv (FlyEM-HB:511051477)')
The neuron variable now holds data about the neuron identified by the given term.
Working with Neuron Data
The retrieved neuron object can provide different representations of neuron data, such as its skeleton, mesh, and volume. These representations can be visualized using various plotting methods.
# Access the skeleton representation
neuron_skeleton = neuron.skeleton
# Check the type of the skeleton representation
print(type(neuron_skeleton)) # Output: <class 'navis.neuron.Neuron'>
# Plot the skeleton in 2D
neuron_skeleton.plot2d()
# Access the mesh representation
neuron_mesh = neuron.mesh
# Access the volume representation
neuron_volume = neuron.volume
Retrieving Multiple Neurons
You can retrieve multiple neurons using the vfb.terms() function, which accepts a list of neuron identifiers.
Example: Retrieving Multiple Neurons
neurons = vfb.terms(['5th s-LNv', 'fru-M-300008', 'catmaid_fafb:8876600'])
This command retrieves multiple neurons, which can then be visualized or manipulated collectively.
Flexible Matching Capabilities
One of the key features of the VFBConnect library is its flexible matching capability. The vfb.terms() function can accept a variety of identifiers, such as:
- IDs: Unique identifiers assigned to each neuron.
- Xref (Cross-references): External references that relate to other datasets.
- Labels: Human-readable names for neurons.
- Symbols: Abbreviated names or symbols used to represent neurons.
- Synonyms: Alternative names by which a neuron might be known.
- Partial Matching: You can provide a partial name, and VFBConnect will attempt to find the best match.
- Case Insensitive Matching: Matching is case insensitive, so if an exact match isn’t found, ‘5th s-LNv’ and ‘5TH S-LNV’ are treated the same. This allows for more flexible querying without worrying about exact case matching.
Example: Using Flexible Matching
neurons = vfb.terms('5th s-LN')
If an exact match isn’t found, VFBConnect will provide potential matches. This feature ensures that even with partial or approximate information, you can still retrieve the relevant neuron data.
Output Example:
Notice: No exact match found, but potential matches starting with '5th s-LN':
'5th s-LNv (FlyEM-HB:511051477)': 'VFB_jrchk8e0',
'5th s-LNv': 'VFB_jrchk8e0'
This notice will help you identify the correct neuron based on the closest matches.
Visualizing Neurons
VFBConnect provides various methods to visualize neuron data, both individually and collectively.
3D Visualization
To plot neurons in 3D, use the plot3d() method. This is useful for visualizing the spatial structure of neurons.
neurons.plot3d()
2D Visualization
For 2D visualization, use the plot2d() method.
neurons.plot2d()
Viewing Merged Templates
VFBConnect also allows viewing merged templates of neurons, combining multiple neuron structures into a single view.
neurons.show()
Opening Neurons in VFB
To open the neurons directly in Virtual Fly Brain, use the open() method. This will launch a browser window displaying the neurons in the VFB interface.
neurons.open()
Summary
- Use
vfb.term()to retrieve single neuron data. - Use
vfb.terms()to retrieve multiple neurons with support for partial, case-insensitive, and flexible matching (IDs, labels, symbols, synonyms, etc.). - Access different data representations (skeleton, mesh, volume) via neuron objects.
- Visualize neuron data in 2D and 3D.
- Use the
show()method to view merged neuron templates. - Open neuron data directly in Virtual Fly Brain with the
open()method.
These examples provide a foundation for working with neuron data from Virtual Fly Brain using the VFBConnect library. By exploring different neuron representations and visualization methods, you can analyze and understand neuron structures more effectively.
3 - Predicted neurotransmitters for EM data now available on VFB
We now incorporate neurotransmitter predictions for Hemibrain neurons from Eckstein et al. (2024) (conf_nt predictions) and MANC neurons from Takemura et al. (2023), with more coming soon.
Note that these studies only worked with a limited set of neurotransmitters and assumed a single neurotransmitter per neuron (see publications for further detail). Neurons with fewer than 100 presynapses are also excluded.
Predictions appear under ‘Relationships’ in the Term Info if available for a particular neuron, giving the confidence level and reference publication.

4 - New Split-GAL4 report pages on FlyBase
FlyBase has added report pages for split driver lines, where two (or more) hemidrivers are combined to restrict functional driver expression to the intersection of the hemidriver expression patterns. These are predominantly ‘split GAL4’ system combinations using a DBD and AD line currently, but more complex combinations may be included in the future.
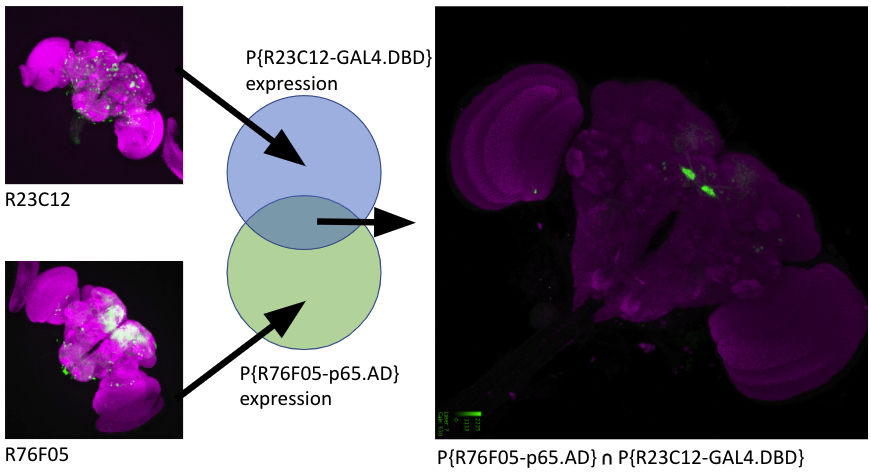
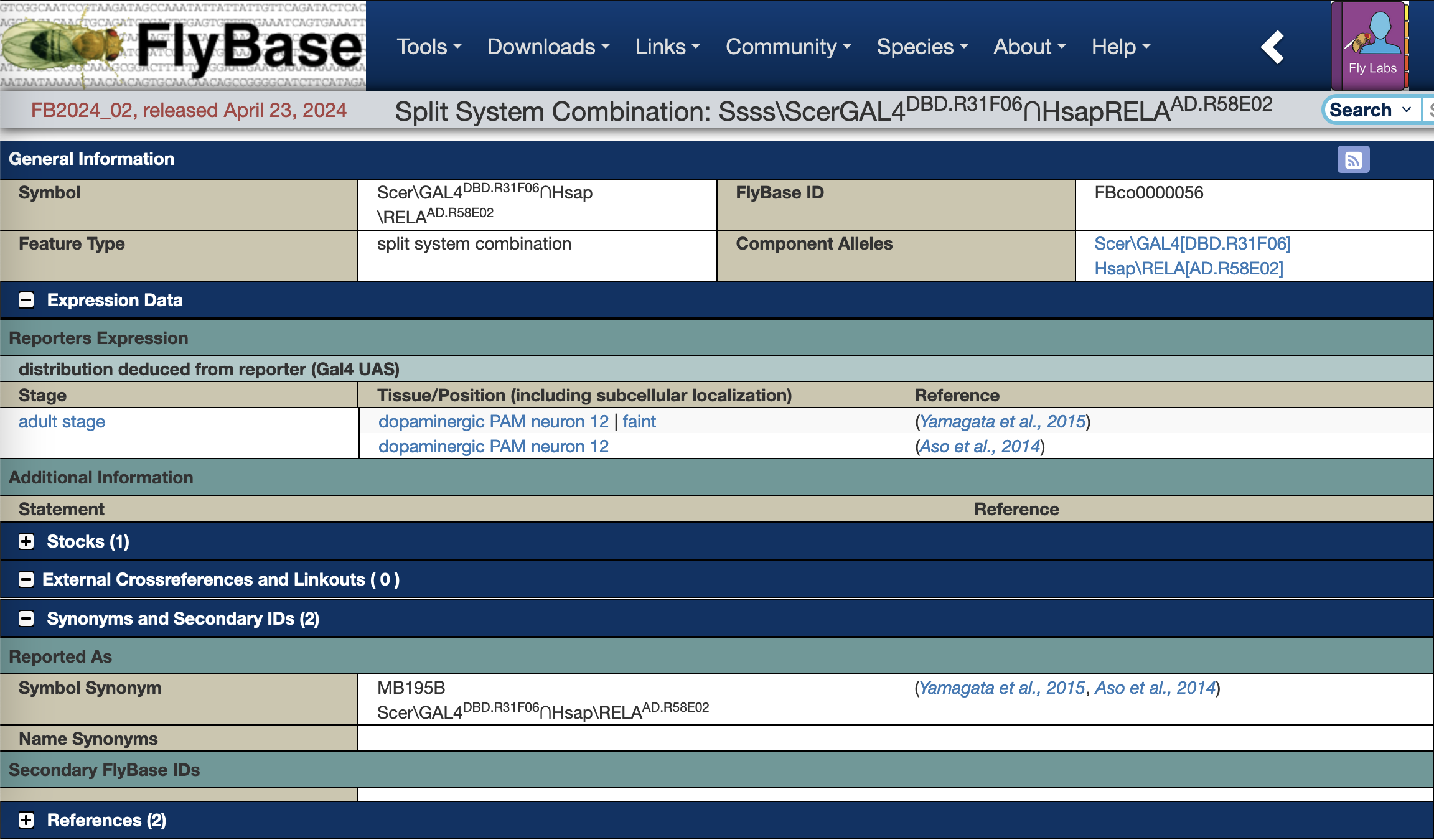
These new ‘split system combinations’ pages (e.g. GAL4DBD.R72B05∩Hsap\RELAAD.ple), include information specific to that combination. Currently these include the FlyBase identifier (FBco), synonyms, the transgenic hemidriver alleles that make up the combination, expression pattern data and links to stocks corresponding to the combination.
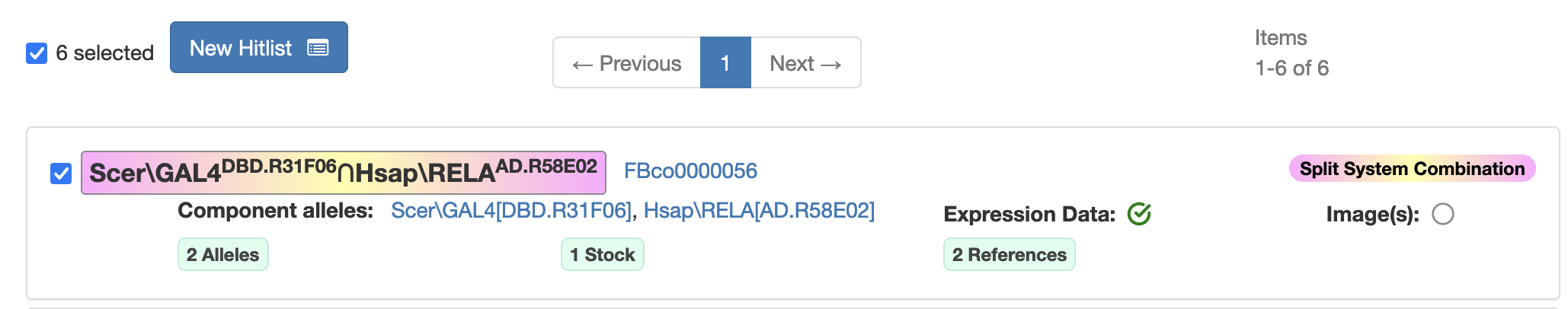
The combination pages have been added to hitlists, so if you search for a regulatory region that drives expression of one of the hemidrivers in a combination (e.g. R72B05, VT064569, ple) you will see the relevant combination(s) in the hitlist. Searching for combination names is also supported (e.g. MB195B). These new report pages will get improvements during the coming months, including images of expression patterns when available.
5 - Transcriptomics data available for cell types on VFB
We now incorporate scRNAseq data from multiple studies, including the Fly Cell Atlas project and other datasets that identify nervous system cell types. These datasets can be found by searching for ‘scRNAseq’ and filtering to ‘Dataset’.
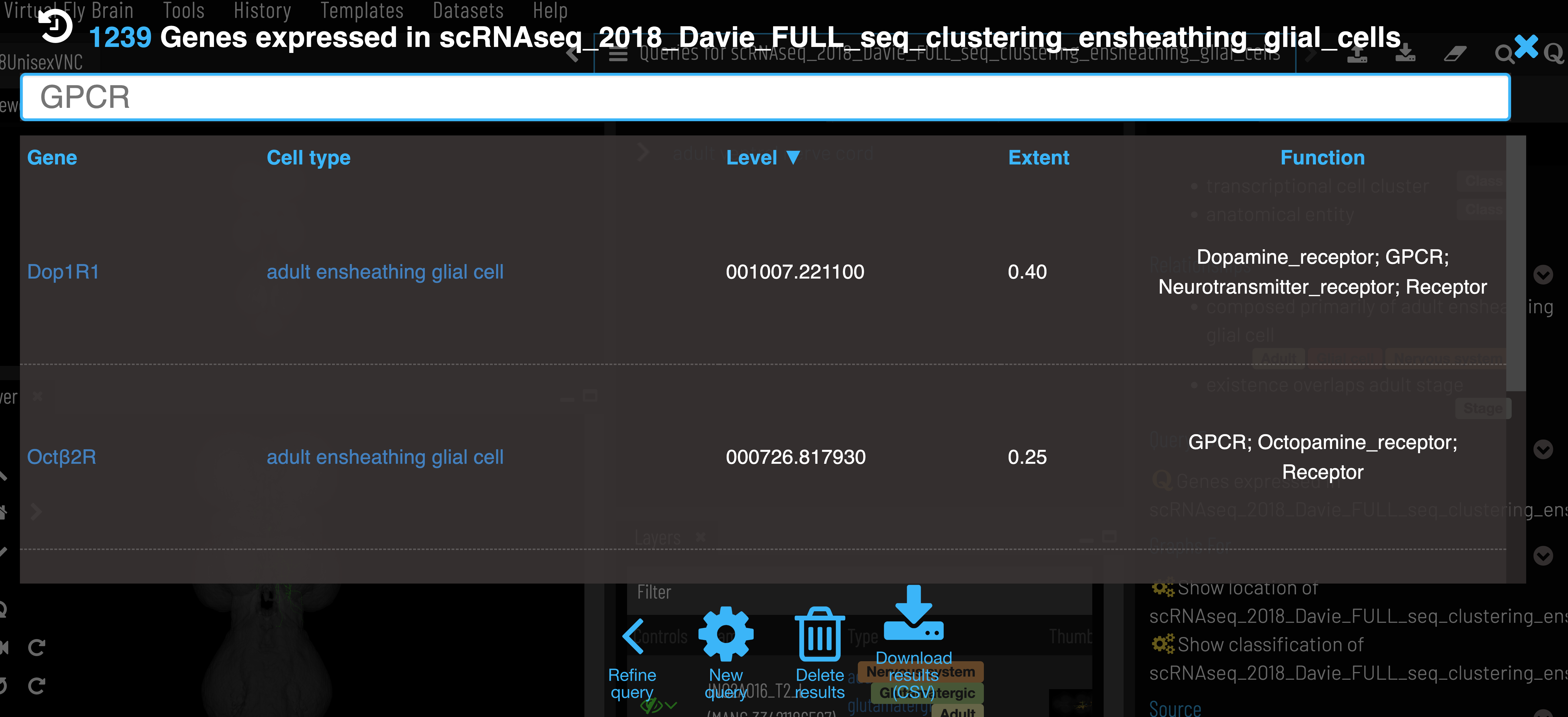
Clusters can be found for particular cell types via the ‘Single cell transcriptomics data for..’ query on the cell type Term Info pane.

Genes for a particular cluster can be filtered by function and sorted by expression level and extent (proportion of cells in cluster expressing the gene). As with other VFB search results, these can be exported as a csv. Note that we currently only include genes that have extent > 0.2 in a cluster.
Data can also be retrieved using VFB_connect.
We pull scRNAseq data from FlyBase, which takes it from the Single Cell Expression Atlas.
6 - Larval split-GAL4 line raw images release
The raw image data for the larval split-GAL4 and Gen 1 LexA lines from Meissner, G. W., et al. “A split-GAL4 driver line resource for Drosophila CNS cell types.” bioRxiv (2024)._ DOI: 10.1101/2024.01.09.574419 is available on VFB here: https://raw.larval.flylight.virtualflybrain.org/
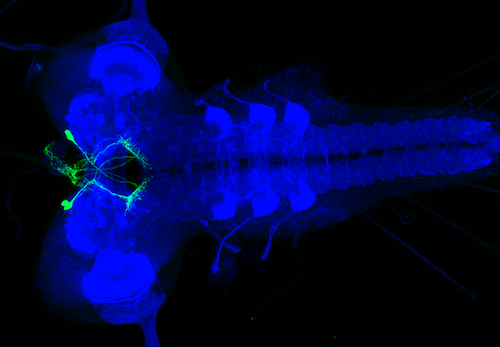
For each image, the confocal stack in TIFF format can be downloaded by selecting an image or all TIFF stacks and z-projections for a line can be downloaded by selecting a line name. Line and image names can be searched at the top right.
We are working on fully integrating these images into VFB. This raw data site does not include full genotypes but these will be included in the curated release on the main VFB site in a later release.
The larval split-GAL4 line release was annotated and provided via the Zlatic lab
7 - Fix for problems with current Safari release
We are aware that VFB’s secure websocket connection is failing to connect on the latest version of Safari on MacOS meaning that no data can be pulled from our servers.
The issue is down to an ’experimental feature’ (NSURLSession WebSocket) being set to ON by default which seams to break most sites using secure websockets.
To turn resolve this issue turn this off:
In Safari on the desktop:
Under 'Develop' expand 'Experimental Features' untick 'NSURLSession WebSocket'
If you don’t see the Develop menu in the menu bar, choose Safari > Preferences, click Advanced, then select “Show Develop menu in menu bar”. Restarting Safari may be necessary if the list of features under ‘Experimental Features’ are greyed out.
On IOS devices:
Open the Settings app
Scroll down and tap on “Safari”
Scroll down to the bottom and tap on “Advanced”
Tap on “Experimental Features” at the bottom
Scroll down until you see “NSURLSession WebSocket"
Disable “NSURLSession WebSocket”
8 - Google search results issue
Google Statement:
December 6-14
An internal issue caused an increase of redirect errors during indexing, and associated email notifications.
This was not due to any website issues, but was due to an internal Google issue.
Pages that were dropped from the index have been scheduled for recrawling.