This is the multi-page printable view of this section. Click here to print.
Archived sites hosted by Virtual Fly Brain
- 1: BrainTrap
- 2: FAFB CATMAID Data Viewer
- 3: FANC CATMAID Data Viewer
- 4: FLYBRAIN Neuron Database (NDB)
- 5: IAV-ROBO CATMAID Data Viewer
- 6: IAV-TNT CATMAID Data Viewer
- 7: L1EM CATMAID Data Viewer
1 - BrainTrap
https://braintrap.virtualflybrain.org/
Overview
BrainTrap was developed as part of a large-scale collaborative research project to collect protein expression location information from protein-trap lines generated by the Cambridge Protein Trap (CPT) project. The database provides detailed 3D confocal datasets showing protein expression patterns throughout the adult Drosophila brain.
Features
- Interactive web-based viewer for exploring 3D confocal datasets
- Full-size images throughout the brain volume can be viewed interactively
- Secondary immunohistochemical label (anti-brp) aids navigation and helps identify brain structures
- Searchable annotations linked to the FlyBase Drosophila anatomy ontology
- Anatomical search criteria can be specified using:
- Automatic term completion
- Hierarchical browser for the ontology
- Annotation provenance tracking with highlighted expression locations
- Downloadable confocal dataset files in original format
- XML format annotation data export
- Gene comparison tables for selected genes
Original Publication
Knowles-Barley S, Longair M and Armstrong JD (2010). BrainTrap: a database of 3D protein expression patterns in the Drosophila brain. Database, Vol. 2010, Article ID baq005, doi:10.1093/database/baq005 (https://doi.org/10.1093/database/baq005)
Funding
Original development was funded by:
- Engineering and Physical Sciences Research Council (EPSRC)
- Medical Research Council
- Biotechnology and Biological Sciences Research Council (BBSRC)
- British Society for Developmental Biology
- Society for Experimental Biology
- Virtual Fly Brain e-Science Institute (ESI) Theme
The fly lines were generated by the Cambridge Protein Trap Project, supported by a Wellcome Trust grant to Kathryn Lilley, Steve Russell and Daniel St. Johnson.
Current Maintenance
This resource is currently archived and maintained by Virtual Fly Brain (VFB).
Current website: https://braintrap.virtualflybrain.org/
Source code repository: https://github.com/VirtualFlyBrain/BrainTrap
Original Development
School of Informatics
The University of Edinburgh
Edinburgh, EH8 9AB, UK
Citation
If you use BrainTrap in your research, please cite:
Knowles-Barley S, Longair M and Armstrong JD (2010). BrainTrap: a database of 3D protein expression patterns in the Drosophila brain. Database, Vol. 2010, Article ID baq005, doi:10.1093/database/baq005 (https://doi.org/10.1093/database/baq005)
2 - FAFB CATMAID Data Viewer
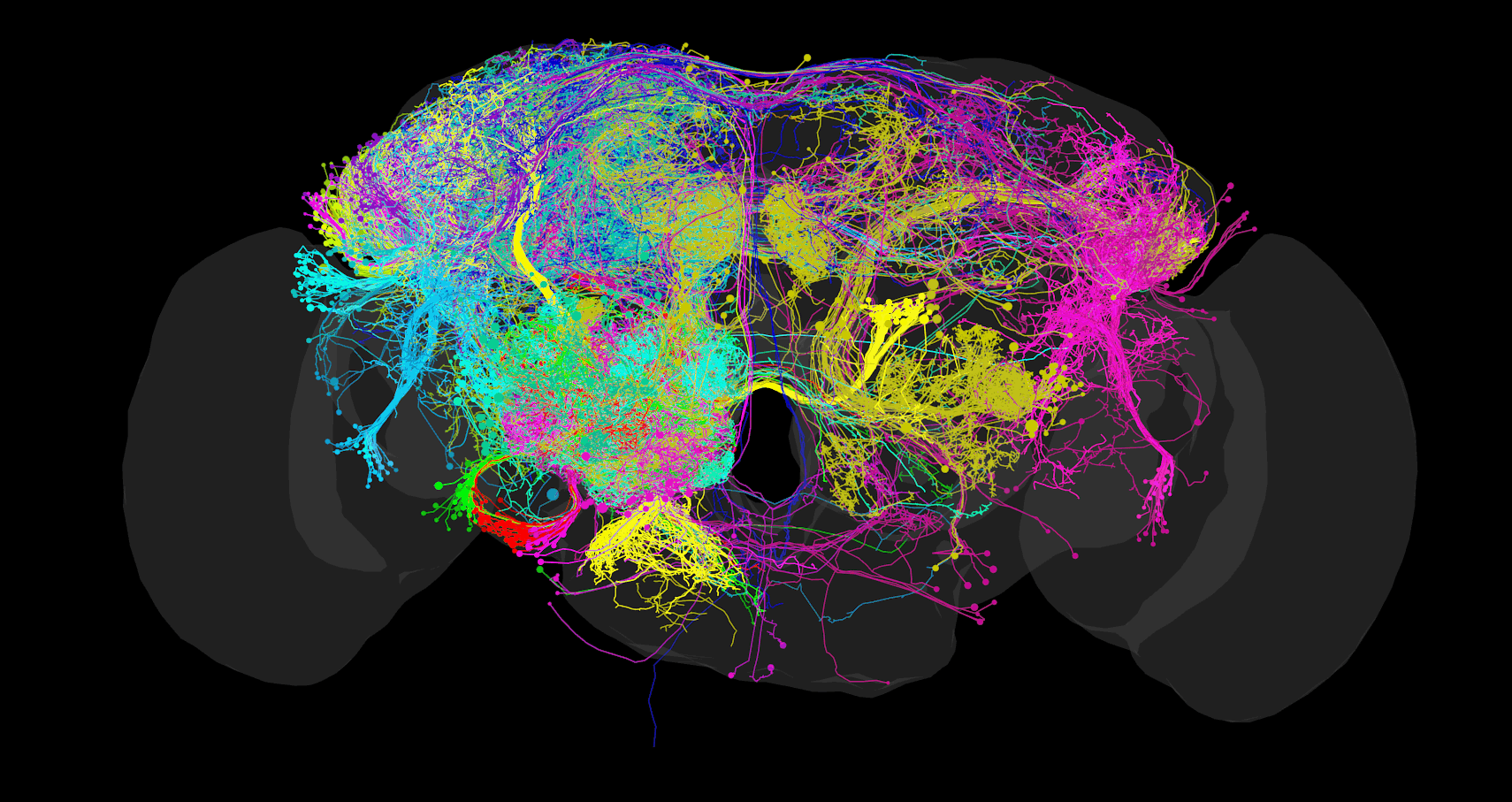
Access
The FAFB CATMAID instance is hosted and maintained by Virtual Fly Brain (VFB) as part of their mission to integrate and preserve key Drosophila neuroscience datasets. The instance is accessible at: https://fafb.catmaid.virtualflybrain.org/
This resource provides access to the FAFB dataset and its associated neural reconstructions. Virtual Fly Brain ensures its long-term availability to the research community.
Source Publications
Primary Resources
- FAFB Project: https://flyconnecto.me/
- EM Dataset: Zheng Z, et al. (2018) A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell, 174(3), 730-743.e22. https://doi.org/10.1016/j.cell.2018.06.019
Contributing Publications
The database includes neurons traced and published in numerous studies. Each neuron is annotated with its source publication. Major contributing publications include:
- Circuit reorganization in the Drosophila mushroom body calyx accompanies memory consolidation (Baltruschat et al. 2021)
- Complete connectome of the Drosophila mushroom body provides insights into memory organization (Bates and Schlegel et al. 2020)
- The Connectome of the Adult Drosophila Mushroom Body Provides Insights into Function (Coates et al. 2020)
- Integration of the olfactory code across dendritic claws of single mushroom body neurons (Dolan and Belliart-Guérin et al. 2018)
- Neurogenetic dissection of the Drosophila lateral horn reveals major outputs, diverse behavioural functions, and interactions with the mushroom body (Dolan et al. 2019)
- Drosophila gustatory projections are segregated by taste modality and connectivity (Engert et al. 2022)
- Re-evaluation of learned information in Drosophila (Felsenberg et al. 2018)
- Motor neurons generate pose-targeted movements via proprioceptive sculpting (Gorko et al. 2024)
- Distinct subpopulations of mechanosensory chordotonal organ neurons elicit grooming of the fruit fly antennae (Hampel et al. 2020)
- A neural circuit for wind-guided olfactory navigation (Kim et al. 2020)
- Parallel encoding of sensory history and behavioral preference during Caenorhabditis elegans olfactory learning (Kind et al. 2021)
- A neural circuit for diurnal temperature preference in Drosophila (Marin et al. 2020)
- Spatial readout of visual looming in the central brain of Drosophila (Morimoto et al. 2020)
- Input Connectivity Reveals Additional Heterogeneity of Dopaminergic Reinforcement in Drosophila (Otto et al. 2020)
- A Neural Circuit Architecture for Angular Integration in Drosophila (Sayin et al. 2019)
- Taste quality and hunger interactions in a feeding sensorimotor circuit (Shiu et al. 2022)
- Generating parallel representations of position and identity in the olfactory system (Taisz et al. 2023)
- A synaptic threshold mechanism for computing escape decisions (Turner-Evans et al. 2020)
- Neural circuitry linking mating and egg laying in Drosophila females (Wang et al. 2020a)
- Circuit and Behavioral Mechanisms of Sexual Rejection by Drosophila Females (Wang et al. 2020b)
- Neural circuit mechanisms of sexual receptivity in Drosophila females (Wang et al. 2020c)
- Zhao et al. (2023)
- Structured sampling of olfactory input by the fly mushroom body (Zheng et al. 2022)
Full references for specific neurons can be found on Virtual Fly Brain by searching for their skeleton ID (skid).
Dataset Contents
The viewer provides access to:
- Complete electron microscopy volume of an adult Drosophila melanogaster brain
- Manually traced neuron reconstructions from multiple research groups
- Synaptic connectivity information
- Associated metadata and annotations
Features
This CATMAID instance enables:
- Browser-based visualization of the serial section electron microscopy data
- Navigation through image stacks
- Viewing of neuron reconstructions
- Analysis of synaptic connectivity
- Data export functionality
Available Tools
- Tracing tool for examining reconstructions
- Neuron search interface with paper-based filtering
- Connectivity analysis tools
- Skeleton visualization options
- API access for programmatic data retrieval (documentation: https://catmaid.readthedocs.io/en/stable/api.html): https://fafb.catmaid.virtualflybrain.org/apis/
Citation Guidelines
When using this data, please cite:
-
The FAFB EM dataset: Zheng Z, et al. (2018) A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell, 174(3), 730-743.e22. https://doi.org/10.1016/j.cell.2018.06.019
-
The CATMAID platform: Saalfeld S, Cardona A, Hartenstein V, Tomančák P (2009) CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25(15): 1984-1986. https://doi.org/10.1093/bioinformatics/btp266
-
The specific publication(s) associated with any neurons you analyze or reference (see Contributing Publications section)
Maintenance & Support
This resource is archived, hosted, and maintained by Virtual Fly Brain (VFB - https://virtualflybrain.org) as part of their commitment to preserving and making accessible critical Drosophila neuroscience data resources.
3 - FANC CATMAID Data Viewer
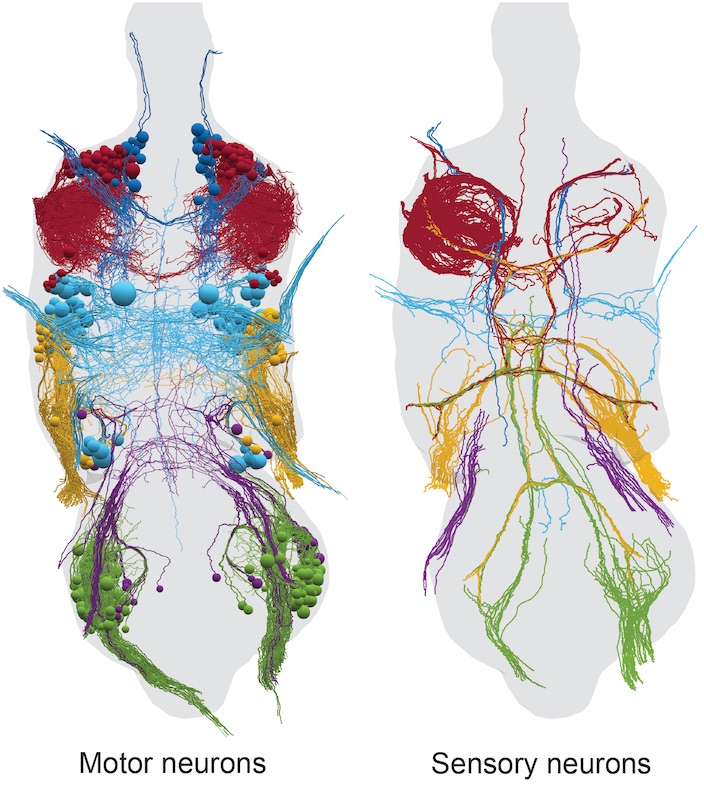
Access
The FANC CATMAID instance is hosted and maintained by Virtual Fly Brain (VFB) as part of their mission to integrate and preserve key Drosophila neuroscience datasets. The instance provides two views of the data:
-
Original EM stack and tracings (Project ID 1): https://fanc.catmaid.virtualflybrain.org/
-
Neurons aligned to JRC2018 VNC female template (Project ID 2): https://fanc.catmaid.virtualflybrain.org/
This resource provides access to both the original FANC dataset and template-aligned reconstructions. Virtual Fly Brain ensures its long-term availability to the research community.
Source Publication
The data is from the research published in:
Fushiki A, et al. (2021) A circuit mechanism for the propagation of waves of muscle contraction in Drosophila. Cell, 184(3), 759-774.e20. https://doi.org/10.1016/j.cell.2020.12.013
see: https://www.lee.hms.harvard.edu/resources
Dataset Contents
The viewer provides access to:
- Complete electron microscopy volume of an adult female Drosophila ventral nerve cord
- Manually traced neuron reconstructions in original EM space
- Template-aligned neurons in JRC2018 VNC female template space
- Synaptic connectivity information
- Associated metadata and annotations
Features
This CATMAID instance enables:
- Browser-based visualization of the serial section electron microscopy data
- Navigation through image stacks
- Viewing of neuron reconstructions in both original and template space
- Analysis of synaptic connectivity
- Data export functionality
Available Tools
- Tracing tool for examining reconstructions
- Neuron search interface
- Connectivity analysis tools
- Skeleton visualization options
- API access for programmatic data retrieval (documentation: https://catmaid.readthedocs.io/en/stable/api.html): https://fanc.catmaid.virtualflybrain.org/apis/
Note: Both views (original and aligned) can be accessed through the same API using different project IDs (pid).
Citation Guidelines
When using this data, please cite:
-
The FANC dataset: Fushiki A, et al. (2021) A circuit mechanism for the propagation of waves of muscle contraction in Drosophila. Cell, 184(3), 759-774.e20. https://doi.org/10.1016/j.cell.2020.12.013
-
The CATMAID platform: Saalfeld S, Cardona A, Hartenstein V, Tomančák P (2009) CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25(15): 1984-1986. https://doi.org/10.1093/bioinformatics/btp266
Maintenance & Support
This resource is archived, hosted, and maintained by Virtual Fly Brain (VFB - https://virtualflybrain.org) as part of their commitment to preserving and making accessible critical Drosophila neuroscience data resources. The aligned neurons are also available through the Virtual Fly Brain website.
4 - FLYBRAIN Neuron Database (NDB)
https://flybrain-ndb.virtualflybrain.org/
Database Structure
FLYBRAIN NDB is a relational database searchable by two main criteria:
1. Neuron Search
Search neurons by:
- Name (full or partial)
- Areas of arborizations
- Distribution of pre- or postsynaptic sites (where known)
- Keywords in documentation
2. Strain/Antibody Search
Search molecular markers including:
- Antibodies
- Drosophila strains (e.g., GAL4 enhancer-trap lines)
- Specific neural pathways
Data Categories
- Visual Neurons
- Olfactory Neurons
- Auditory Neurons
- Gustatory Neurons
- Central Complex Neurons
- Clonal Units
- Brain Region Definitions
- GAL4 and LexAV Strain Collections
Brain Explorer Features
The online brain browsing system allows users to:
- Rotate volume-rendered images
- Create sections
- Adjust magnification
- Browse confocal data
Terms of Use
Access
- FLYBRAIN NDB is freely available
- No login or registration required
Copyright
- All contents © 2008 FLYBRAIN NDB or their original publication
- Copyright is noted in respective images
Usage Rights
- Free use for non-profit, educational, and scientific purposes with proper citation
- Republication or commercial use requires consent from the FLYBRAIN NDB manager
Citation Guidelines
Citing the Database
When citing the database as a whole, use: “Flybrain Neuron Database (https://flybrain-ndb.VirtualFlyBrain.org)”
Citing Specific Records
Format: [Neuron type], [Author(s)], ([Year]) Flybrain Neuron Database (https://flybrain-ndb.VirtualFlyBrain.org), acc. [number]
Example: “LC10 neuron, Otsuna, H., and Ito, K. (2007) Flybrain Neuron Database (https://flybrain-ndb.VirtualFlyBrain.org), acc. 10010”
Important Notes:
- Include accession numbers when citing specific neurons
- Cite original publications where applicable
- Multiple contributors may exist for each record
Current Maintenance
This resource is currently archived and maintained by Virtual Fly Brain (VFB).
Source code repository: https://github.com/VirtualFlyBrain/flybrain-ndb
Original Development
© 2010 FLYBRAIN Neuron Database
Original Contact: Kazunori Shinomiya
5 - IAV-ROBO CATMAID Data Viewer
Access
The IAV-ROBO CATMAID instance is hosted and maintained by Virtual Fly Brain (VFB) as part of their mission to integrate and preserve key Drosophila neuroscience datasets. The instance is accessible at: https://iav-robo.catmaid.virtualflybrain.org/
This resource provides a direct view into the dataset used to study the organization of proprioceptive circuits in Drosophila larvae. Virtual Fly Brain ensures its long-term availability to the research community.
Source Publication
This data is from the research published in:
Valdes-Aleman J, et al. (2021). Comparative Connectomics Reveals How Partner Identity, Location, and Activity Specify Synaptic Connectivity in the Drosophila Motor System. Neuron, 109(1), 105-120.e7. https://doi.org/10.1016/j.neuron.2020.10.004
Dataset Contents
The viewer provides access to:
- Serial section electron microscopy data of Drosophila 1st instar larval CNS (1.5-segment fraction, A1/A2 segment)
- Image resolution: 3.8 nm x 3.8 nm x 40 nm (x, y, z)
- Genotype: w;; iav-GAL4/UAS-FraRobo
- Manually traced neuron reconstructions showing shifted mechanosensory projections
- Synaptic connectivity information
- Associated metadata and annotations
Technical Details
The EM volume was prepared and imaged following protocols detailed in Ohyama et al. (2015). Neurons were reconstructed using CATMAID to obtain skeletonized structures and connectivity information. All synaptic connections represent chemical synapses.
Despite the FraRobo-induced lateral shift in terminal projections, neurons remain uniquely identifiable based on:
- Nerve entry point of the main neurite into the neuropil
- Growth pattern of main axonal and dendritic branches
- Bilateral vs ipsilateral projections
For example, mechanosensory chordotonal neurons can be identified by their:
- Eight stereotypic entry points per hemisegment
- Most lateral position among sensory neurons in their nerve bundle
- Further lateral shift due to FraRobo expression
Features
This CATMAID instance enables:
- Browser-based visualization of the serial section electron microscopy data
- Navigation through image stacks
- Viewing of neuron reconstructions
- Analysis of synaptic connectivity
- Data export functionality
Available Tools
- Tracing tool for examining reconstructions
- Neuron search interface
- Connectivity analysis tools
- Skeleton visualization options
- API access for programmatic data retrieval (documentation: https://catmaid.readthedocs.io/en/stable/api.html): https://iav-robo.catmaid.virtualflybrain.org/apis/
Citation Guidelines
When using this data, please cite both:
-
The original research: Valdes-Aleman J, et al. (2021). Comparative Connectomics Reveals How Partner Identity, Location, and Activity Specify Synaptic Connectivity in the Drosophila Motor System. Neuron, 109(1), 105-120.e7. https://doi.org/10.1016/j.neuron.2020.10.004
-
The CATMAID platform: Saalfeld S, Cardona A, Hartenstein V, Tomančák P (2009) CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25(15): 1984-1986. https://doi.org/10.1093/bioinformatics/btp266
Maintenance & Support
This resource is archived, hosted, and maintained by Virtual Fly Brain (VFB - https://virtualflybrain.org) as part of their commitment to preserving and making accessible critical Drosophila neuroscience data resources.
6 - IAV-TNT CATMAID Data Viewer
Access
The IAV-TNT CATMAID instance is hosted and maintained by Virtual Fly Brain (VFB) as part of their mission to integrate and preserve key Drosophila neuroscience datasets. The instance is accessible at: https://iav-tnt.catmaid.virtualflybrain.org/
This resource provides a direct view into the dataset used to study the organization of proprioceptive circuits in Drosophila larvae. Virtual Fly Brain ensures its long-term availability to the research community.
Source Publication
This data is from the research published in:
Valdes-Aleman J, et al. (2021). Comparative Connectomics Reveals How Partner Identity, Location, and Activity Specify Synaptic Connectivity in the Drosophila Motor System. Neuron, 109(1), 105-120.e7. https://doi.org/10.1016/j.neuron.2020.10.004
Dataset Contents
The viewer provides access to:
- Serial section electron microscopy data of Drosophila 1st instar larval CNS (Control 1: whole-CNS volume, A1 segment)
- Image resolution: 3.8 nm x 3.8 nm x 40 nm (x, y, z)
- Manually traced neuron reconstructions
- Synaptic connectivity information
- Associated metadata and annotations
Technical Details
The EM volume was prepared and imaged following protocols detailed in Ohyama et al. (2015). Neurons were reconstructed using CATMAID to obtain skeletonized structures and connectivity information. All synaptic connections represent chemical synapses.
The neurons are uniquely identifiable based on key morphological features:
- Nerve entry point of the main neurite into the neuropil
- Growth pattern of main axonal and dendritic branches
- Bilateral vs ipsilateral projections
- Position of terminal projections within the medio-lateral, dorso-ventral and antero-posterior axes
Features
This CATMAID instance enables:
- Browser-based visualization of the serial section electron microscopy data
- Navigation through image stacks
- Viewing of neuron reconstructions
- Analysis of synaptic connectivity
- Data export functionality
Available Tools
- Tracing tool for examining reconstructions
- Neuron search interface
- Connectivity analysis tools
- Skeleton visualization options
- API access for programmatic data retrieval (documentation: https://catmaid.readthedocs.io/en/stable/api.html): https://iav-tnt.catmaid.virtualflybrain.org/apis/
Citation Guidelines
When using this data, please cite both:
-
The original research: Valdes-Aleman J, et al. (2021). Comparative Connectomics Reveals How Partner Identity, Location, and Activity Specify Synaptic Connectivity in the Drosophila Motor System. Neuron, 109(1), 105-120.e7. https://doi.org/10.1016/j.neuron.2020.10.004
-
The CATMAID platform: Saalfeld S, Cardona A, Hartenstein V, Tomančák P (2009) CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25(15): 1984-1986. https://doi.org/10.1093/bioinformatics/btp266
Maintenance & Support
This resource is archived, hosted, and maintained by Virtual Fly Brain (VFB - https://virtualflybrain.org) as part of their commitment to preserving and making accessible critical Drosophila neuroscience data resources.
7 - L1EM CATMAID Data Viewer
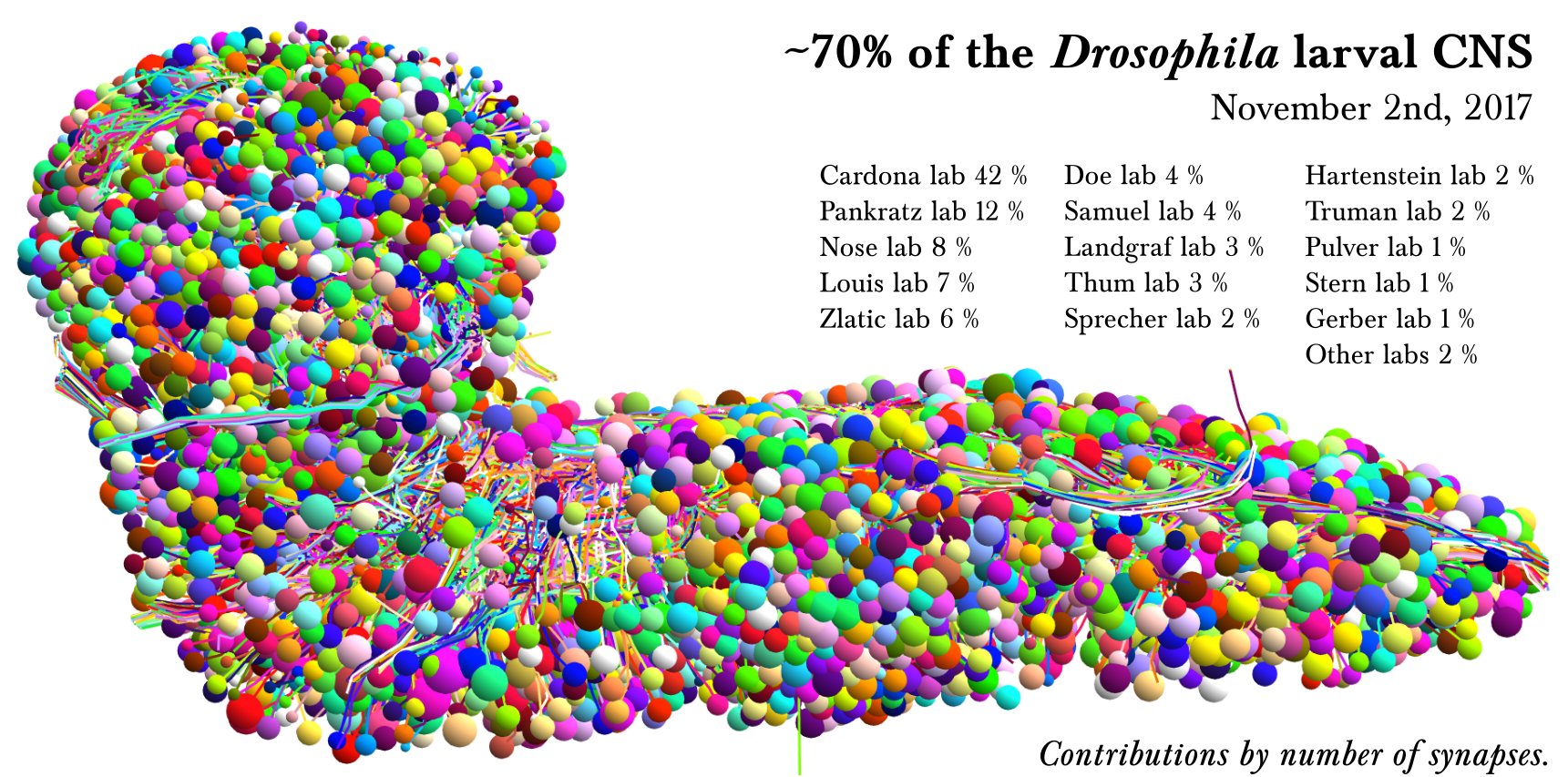
Access
The L1EM CATMAID instance is hosted and maintained by Virtual Fly Brain (VFB) as part of their mission to integrate and preserve key Drosophila neuroscience datasets. The instance is accessible at: https://l1em.catmaid.virtualflybrain.org/
This resource provides access to the L1EM dataset and its associated neural reconstructions. Virtual Fly Brain ensures its long-term availability to the research community.
Source Publication
Primary Resource
- L1EM Dataset: Winding M, et al. (2023) The connectome of an insect brain. Science, 379(6636):eadd9330. https://doi.org/10.1126/science.add9330
Contributing Publications
The database includes neurons traced and published in numerous studies. Each neuron is annotated with its source publication. Major contributing publications include:
- Andrade et al. (2019)
- Barnes et al. (2022)
- Berck, Khandelwal et al. (2016)
- Burgos et al. (2018)
- Carreira-Rosario, Arzan Zarin, Clark et al. (2018)
- Eichler, Li, Litwin-Kumar et al. (2017)
- Eschbach, Fushiki et al. (2020, 2020b)
- Fushiki et al. (2016)
- Gerhard et al. (2017)
- Heckscher et al. (2015)
- Hueckesfeld et al. (2020)
- Imambocus et al.
- Jovanic et al. (2019)
- Jovanic, Schneider-Mizell et al. (2016)
- Larderet, Fritsch et al. (2017)
- Mark et al. (2019)
- Miroschnikow et al. (2018)
- Ohyama, Schneider-Mizell et al. (2015)
- Schlegel et al. (2016)
- Takagi et al. (2017)
- Tastekin et al. (2018)
- Valdes-Aleman et al. (2021)
- Winding, Pedigo et al. (2023)
- Zarin, Mark et al. (2019)
- Zwart et al. (2016)
Full references for specific neurons can be found on Virtual Fly Brain by searching for their skeleton ID (skid).
Dataset Contents
The viewer provides access to:
- Complete electron microscopy volume of a Drosophila first instar larva brain
- Manually traced neuron reconstructions from multiple research groups
- Synaptic connectivity information
- Associated metadata and annotations
Features
This CATMAID instance enables:
- Browser-based visualization of the serial section electron microscopy data
- Navigation through image stacks
- Viewing of neuron reconstructions
- Analysis of synaptic connectivity
- Data export functionality
Available Tools
- Tracing tool for examining reconstructions
- Neuron search interface with paper-based filtering
- Connectivity analysis tools
- Skeleton visualization options
- API access for programmatic data retrieval (documentation: https://catmaid.readthedocs.io/en/stable/api.html): https://l1em.catmaid.virtualflybrain.org/apis/
Citation Guidelines
When using this data, please cite:
-
The L1EM dataset: Winding M, et al. (2023) The connectome of an insect brain. Science, 379(6636):eadd9330. https://doi.org/10.1126/science.add9330
-
The CATMAID platform: Saalfeld S, Cardona A, Hartenstein V, Tomančák P (2009) CATMAID: collaborative annotation toolkit for massive amounts of image data. Bioinformatics 25(15): 1984-1986. https://doi.org/10.1093/bioinformatics/btp266
-
The specific publication(s) associated with any neurons you analyze or reference (see Contributing Publications section)
Maintenance & Support
This resource is archived, hosted, and maintained by Virtual Fly Brain (VFB - https://virtualflybrain.org) as part of their commitment to preserving and making accessible critical Drosophila neuroscience data resources.